
ページ読込中...

ページ読込中...

[English]
| 安田 賢二 [教授] |  |
|
| homepage | https://www.yasuda.phys.waseda.ac.jp | |
| 専門分野 | 生物物理学 | |
| 研究テーマ・研究活動 | ||
|
||
生物は、分子から細胞、個体に至る階層構造を持ったシステムとして機能しています。単純な原子、分子の集合化のみでは構築できない「生命」の特徴を生み出している本質はどこにあるのでしょうか。また、この「生命」の神秘はすべて物理法則のみで記述し、説明できるのでしょうか。
物理学の世界では原子、分子が規則性を持った現象論で議論をするに十分な振る舞いを見せるためにはアボガドロ数程度の要素の集合が必要ですが、生命システムの中ではわずか数個の要素を組み合わせることで巧みに高い規則性、再現性、保存性を維持することができます。これは、まさに生命システムが持つ「集団効果」であり、これを実現するための規則性、秩序をもたらす巧みな機構が生命の本質の一つなのかもしれません。
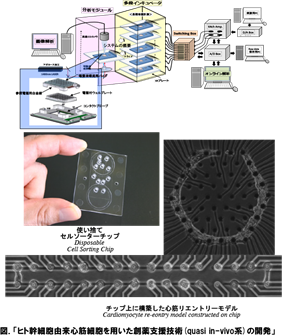 わたしたちは、この生命の各階層での要素1単位とその集団効果について、構成的な手法を用いて解明を進めています。たとえば、化学・力学エネルギー変換システムである生体分子モーターについては、その1分子の機構の解明と、これが自己組織化した筋原線維の集団的協同(振動)現象を、細胞レベルでは、心筋細胞や神経細胞1細胞の振る舞いと、これが空間配置を持って集団化したときの振る舞いの比較解析を行い、そこから「臓器モデル」ともいえる最小の細胞ネットワークチップなども構築に成功しており、創薬スクリーニングでの応用も始まりつつあります。このように、生物物理学の生物学との最大の違いは、物理法則の理解の上に立った生命現象の理解なのですが、もう一つの特徴 として、理解するために必要な技術も自分たちで創り上げています。
わたしたちは、この生命の各階層での要素1単位とその集団効果について、構成的な手法を用いて解明を進めています。たとえば、化学・力学エネルギー変換システムである生体分子モーターについては、その1分子の機構の解明と、これが自己組織化した筋原線維の集団的協同(振動)現象を、細胞レベルでは、心筋細胞や神経細胞1細胞の振る舞いと、これが空間配置を持って集団化したときの振る舞いの比較解析を行い、そこから「臓器モデル」ともいえる最小の細胞ネットワークチップなども構築に成功しており、創薬スクリーニングでの応用も始まりつつあります。このように、生物物理学の生物学との最大の違いは、物理法則の理解の上に立った生命現象の理解なのですが、もう一つの特徴 として、理解するために必要な技術も自分たちで創り上げています。
| Kenji Yasuda [Professor] |  |
|
| homepage | https://www.yasuda.phys.waseda.ac.jp | |
| research field | Biophysics | |
| research keywords | ||
|
||
| link | ||
The cells in a group are individual entities, and differences arise even among cells with identical genetic information that have grown under the same conditions. These cells respond differently to perturbations. Why and how do these differences arise? Cells are minimum units determining their responses through genetic and epigenetic information like the history of interactions between them and fluctuations in environmental conditions affecting them. To understand the rules underlying possible differences occurring in cells, we need to develop methods of simultaneously evaluating both the genetic and epigenetic information. In other words, if we are to understand adaptation processes, community effects, and the meaning of cell network patterns, we need to analyze their epigenetic information. We thus started a project focusing on developing a system that could be used to evaluate the epigenetic information in cells by continuously observing specific examples and their interactions under controlled conditions. The importance of understanding epigenetic information is expected to become apparent in cell-based biological and medical fields like cell-based drug screening and the regeneration of organs from stem cells, fields where phenomena cannot be interpreted without taking epigenetic factors into account.
We have started a study on the “determination of genetic and epigenetic control processes in cells” using on-chip microfablication techniques and cell-based analysis. To understand the meaning of genetic information and epigenetic correlation in cells, we developed an on-chip single-cell-based microcultivation method. As we can see in Fig. 1, the strategy behind our method is constructive, involving three steps. First, we purify cells from tissue one by one in a nondestructive manner. We then cultivate and observe them under fully controlled conditions (e.g., cell population, network patterns, or nutrient conditions) using an on-chip single-cell cultivation chip or an on-chip agarose microchamber system. Finally, we do single-cell-based expression analysis through photothermal denaturation and single-molecule level analysis. In this way, we can control the spatial distribution and interactions of cells.
The first aim of our single-cell-based study was to develop methods and systems that enable the mechanism responsible for controlling (regulating) cells epigenetically to be analyzed. It is based on the idea that, although genetic information creates a network of biochemical reactions, its history as a parallel-processing recurrent network was ultimately determined by the environmental conditions of cells, which we call epigenetic information. As previously discussed, if we are to understand the events in living systems at the cellular level, we need to bear in mind that epigenetic information complements the genetic information.
The advantage of this approach is that it removes the complexity in underlying physicochemical reactions that are not always completely understood and for which most of the necessary variables cannot be measured. Moreover, this approach shifts the view of cell regulatory processes from a basic chemical ground to a paradigm of the cell as an information-processing unit working as an intelligent machine capable of adaptating to changing environmental and internal conditions. This is an alternative representation of the cell and can bring new insights into cellular processes. Thus, models derived from such a viewpoint can directly help in more traditional biochemical and molecular biological analyses that assist in our understanding of control in cells.
The main purpose of our study is to develop on-chip single-cell-based cultivation and analysis systems to monitor dynamic processes in the cell. We have used these systems to extend ideas from the genetic to the genetic-epigenetic network in investigating topics like variations in cells with the same genetic information, inheritance of non-genetic information between adjacent generations of cells, cellular adaptation processes caused by environmental change, the community effect of cells, and network pattern formation in cell groups (Fig. 2). After sufficient experimental observations, we can understand the role of epigenetic information in modeling more complex signaling cascades. This field has almost been entirely monopolized by physico-chemical models, which provide a good standard for comparison, evaluation, and development with our approach. The ultimate aim of our study is to provide a comprehensive understanding of living systems as products of both genetic and epigenetic information. It would permit us to describe the phenomena occurring in cell systems sufficiently well to be able to interpret and control them.